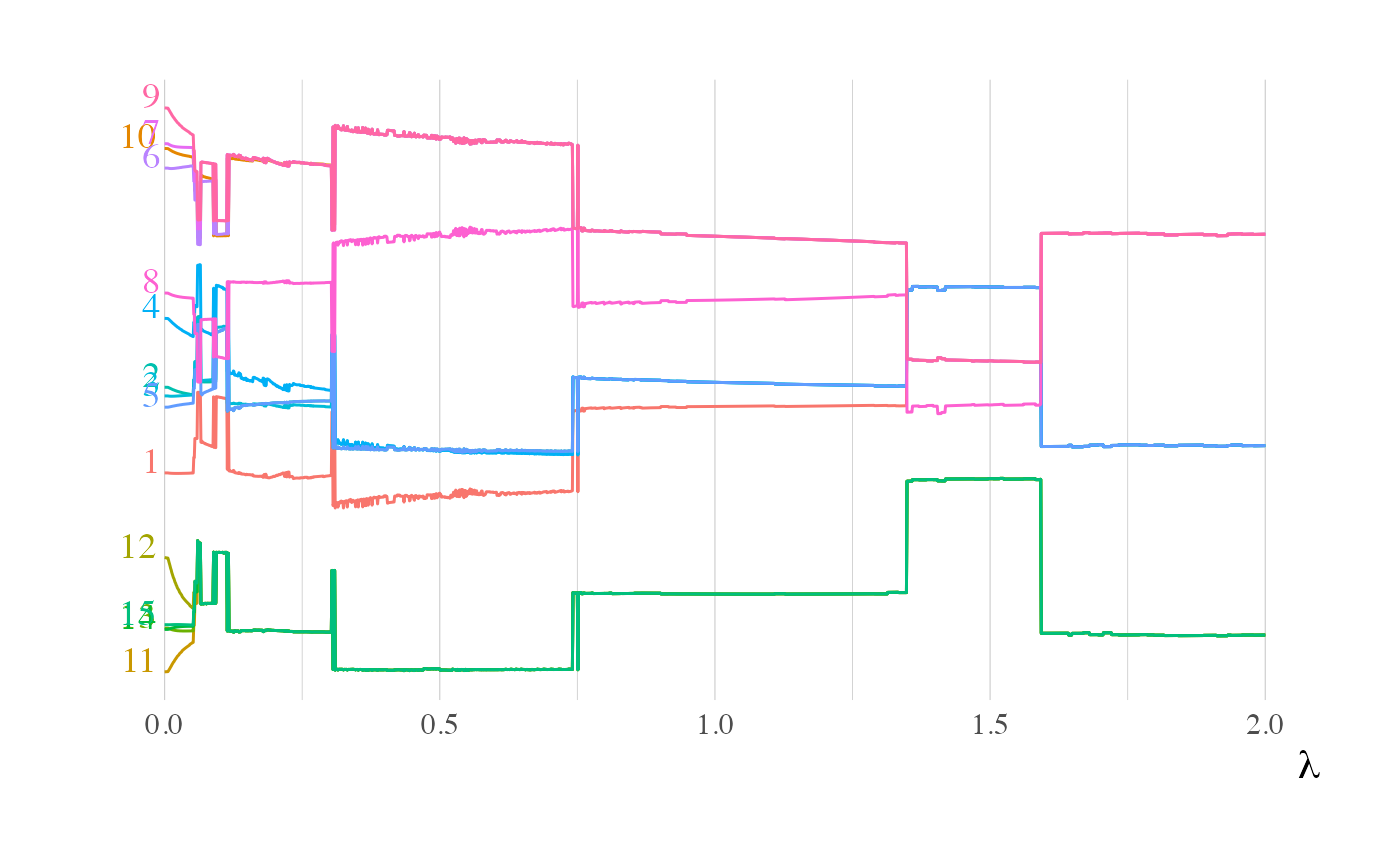
Multidimensional scaling for Clusterpath.
ggdistance.RdVisual representation of the distance evolution between the clusters along \(\lambda\). We recall that for a precision matrix \(\Theta\) which have a block matrix structure, the distance between two variables is defined by: $$ D(\Theta_{i\cdot}, \Theta_{j\cdot}) = \sqrt{\sum_{k\neq i,j} (\Theta_{ik} - \Theta_{jk})^2}, $$ With this distance, two variables in the same cluster have a null distance.
Arguments
- list_results
A list of results optimization from
HR_Clusterpath().- names
A list of name : the names of the variables, if
NULLit will be \(1,...,d\).
Method used for the representation
The cluster's distance is a measure of dissimilarities between variable. With this in mind, we have a dissimilarities matrix \(W\) and we want to build for each \(\lambda\) a reconstition of a 1-dimensional scatter plot with the respect of these dissimilarities.
We use the function cmdscale() from stats package to operate the optimization. It follows
the analysis of \([1]\).
Examples
# Construction of clusters and R matrix
R <- matrix(c(1, -3, 0,
-3, 2, -2,
0, -2, 1), nc = 3)
clusters <- list(1:5, 6:10, 11:15)
# Construction of induced theta and corresponding variogram gamma
Theta <- build_theta(R, clusters)
Gamma <- graphicalExtremes::Theta2Gamma(Theta)
gr3_bal_sim_param_cluster <-
list(
R = R,
clusters = clusters,
Theta = Theta,
Gamma = Gamma,
chi = 1,
n = 1e3,
d = 15
)
set.seed(804)
data <- graphicalExtremes::rmpareto(n = gr3_bal_sim_param_cluster$n,
model = "HR",
par = gr3_bal_sim_param_cluster$Gamma)
lambda <- seq(0, 2, 1e-3)
res <- HR_Clusterpath(data = data,
zeta = gr3_bal_sim_param_cluster$chi,
lambda = lambda,
eps_f = 1e-1)
ggdistance(res)